The task is a quantum of workflow
This post describes how we organize our work over ten years, twenty analysts, dozens of countries, and hundreds of projects: we start with a task. A task is a single chunk of work, a quantum of workflow. Each task is self-contained and self-documenting; I’ll talk about these ideas at length below. We try to keep each task as small as possible, which makes it easy to understand what the task is doing, and how to test whether the results are correct.
In the example I’ll describe here, I’m going to describe work from our Syria database matching project, which includes about 100 tasks. I’ll start with the first thing we do with files we receive from our partners at the Syrian Network for Human Rights (SNHR).
Why so much structure?
Before diving into a task, why are we bothering with all this? If you just want to trust me that it’s worth it, skip to the next section below.
The short answer is that without a lot of structure, we forget what we’ve done in the past, we can’t read each others’ work, and we can’t test whether what we’ve done is correct.
Unix is built on the axiom that “small pieces loosely joined” leads to the fewest bugs and the greatest flexibility. The idea is that each component of work we need to do should be as small and simple as possible. To accomplish a larger task, we link one piece to another. Keeping pieces small makes them simpler to design and debug, and using simple interfaces enables us to connect pieces together to build bigger structures.
We process a lot of data at HRDAG. We get raw datasets which need to be transferred from proprietary formats into formats our open source tools can use; we standardize field values; remove extraneous pieces; fix broken dates; canonicalize names; create unique keys; match and merge databases; and estimate missing field values and missing records, among many, many other tasks. As we add more analysts, more datasets, and more results, it can be increasingly difficult to make sure all the pieces fit together. How can you know if the results are correct? If we fix a bug, or the raw data is updated, how can we recalculate the full results?
Furthermore, we’ve been doing this work for 25 years; we have data and analysis on about 30 countries. In some of the countries, we’ve worked on numerous projects over many years (we’ve been working on Guatemala for 22 years!). In order to keep all this work coherent, we have imposed a standard organization that each project follows.
Whether you need structure like this in your work depends on the scale of your projects. We often say you need this approach when you start accumulating “more-than-two’s:” do you have more than two analysts working together? Will the analysis be used in more than two reports (or for more than two campaigns, or for more than two clients)? Are you using more than two datasets? More than two programming languages? Will the project last more than two years? And so forth. As the project’s complexity grows, the value of a standardized scheme increases.
If a project is organized in a standard way, it’s easier to figure out what’s going on when you drop into it.
A concrete example
We have received a number of data files from SNHR. Each file is a list of individual deaths in an Excel spreadsheet. Each row of each spreadsheet identifies a single death, with columns for the victim’s names, age, location and date of death, professional affiliation, and other information (in Arabic). Our first task is to extract the information from each Excel sheet, and then to append the information from all the various sheets into a single CSV file. This is pretty simple using pandas’s read_excel() and concat() functions, so I’m not going to discuss the mechanics, just the workflow.
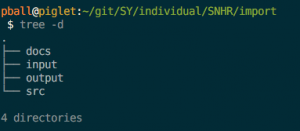
This task is called import. Like every task, it has at minimum three directories: input, src, and output. Here’s how it works:
- Files the task will read are in input/;
- Code that executes to read input files and write output files is in src/;
- Files the task will write are in output/.
The directory structure for the import task can be seen in Figure 1 (see below for more info on the directories).
Files in input/ are read-only. They come from outside the task, and once they’re placed in input, they are never changed.
Source code reads the input files to make output files, and the code is kept in the src/ directory.
Results created by source code are written in output/. These may be log files, transformed data files (csv, hdf5, or feather), graphs (as png or pdf files), tables (in tex files), individual statistics we’ll use in later steps (in yaml files), or anything else our code generates.
Tasks are self-contained and self-documenting
By organizing a task in these three directories, the task becomes self-contained, and even more importantly, self-documenting. Before diving into these two ideas, consider that the simplest reason to use this format is that we never accidentally overwrite files. If we are always reading from input/ and writing to output/, we can’t overwrite our input files.
This scheme also makes it simple to regenerate results: delete everything in the output/ directory, and rerun the code in src/. Just by themselves, these two benefits can help newbie analysts avoid costly mistakes. But on to the bigger, longer-term benefits.
Self-contained
Self-contained means that everything we need to understand how results appear in output/ can be found inside the task. All the data needed is in input/: and this means all the data that is needed. Data might come from lots of different origins: data comes from our partners like SNHR, of course, but more commonly, input data is the output from a prior (“upstream”) task. And all the files to be read by src/ must be in input/ (data from prior tasks can be used via symbolic links with relative paths in input/).
All the transformations are executed — and documented — exclusively by code that can be found in src/ (see below for a few additional directories that are occasionally used). And all the results are in output/, which means that if you want to know what the task creates, the exhaustive answer can always be found just by looking at the output/ directory.
Self-documenting
Self-documentation is the heart of the HRDAG data processing practice. One layer of self-documentation results from being self-contained: if you are reviewing a task, everything you need to know about the task can be seen in the files that are in the task.
In HRDAG’s workflow, the only way we produce a result is by executing code. That means we don’t use interactive tools like spreadsheets or GUI-based statistics software (occasional exceptions are described below).
There are a lot of reasons we require that all results be scripted. For this analysis, there are two crucial points: first, the scripts are the documentation for the results. If the only results we use come from executing code, then the code is the ultimate documentation of the results.
Furthermore, the role of a given file, whether it is an input file, an output file, or an executable file, is indicated by the directory it is in. And the name of the task should clearly express what the task is trying to accomplish.
In the example here, the task’s name is import/, and that makes clear that the data in input/ come directly from the partners, from outside our work. Next, the project path to the task is SY/individual/SNHR/import/, which tells a fuller story: this task belongs to the Syria project, it is part of the processing for an individual dataset (i.e., not merging several datasets together), and the dataset being processed is SNHR. Thus the task’s full path, and the task’s component directories all contribute to expressing the role of each part of the project.
Documentation versus self-documenting projects
A last note on documentation vs self-documentation. Data processing code tends to change rapidly while we’re figuring out how to get a result, but once we’ve got the result, the code stops changing. People who don’t write much code often think that it’s important to write human-language documentation that describes what the code does. I think this is misguided and creates more confusion than clarity.
The problem is that the code changes faster than the documentation. When you return to your work, months later, you can’t figure out why the documentation doesn’t make sense. The answer is that you probably changed the code without updating the documentation. Nothing breaks when you run the code, so you forget about the docs.
There are plenty of kinds of projects that greatly benefit from documentation. For example, library code written by a development team and used by lots of other people needs clear docs; the scikit-learn docs are exemplary. However, documentation in data processing tasks tends to be misleading.
We build the documentation into the structure of the task. If the task works, the structure is right, and that means the self-documentation is in place. This approach doesn’t work for all projects, but we’ve found that for data processing projects, self-documentation — that is, explanatory structures — make reviewing a long-ago, and long-forgotten project relatively easy.
Additional, optional directories
These are rare, but it’s important to keep the meaning of the three core directories clear, so files that don’t fit the input-src-output logic are kept separately.
note: Increasingly we prototype ideas in interactive jupyter notebooks and RStudio markdown files. Notebooks and R markdown files are a great place to explore analyses, and to make sure a tricky chunk of code is doing what you expect. However, they’re not good for the final results. In particular, it’s difficult to assure that the cells of an interactive notebook have been executed in order. Users often test the results in a given cell, then go above that cell to adjust something, then re-run other cells. The results of a notebook may become dependent on an arbitrary execution sequence, which makes them fragile. There are other issues with using notebooks for production work, but from the perspective of the task, execution context and sequence are the biggest problems. We store notebooks in the note/ directory, but the final work has to be in scripts in src/.
docs: Some tasks have additional files we need to keep that aren’t data or source code. For example, when we receive data files from a partner to be stored in an import/ task, we sometimes get documentation from them in pdf, docx, html, or some other format. These files are stored in a doc/ directory at the same level as input/.
hand: In other tasks, we may need to recode a series of values. This is common in data cleaning, for example, where we might want to convert city names “San Fran,” “Babylon by the Bay,” and “Frisco,” to a canonical “San Francisco.” In our practice, we would store the variant spellings in a small csv file which we maintain by hand, like source code. It’s data, not code, but it’s not an input because we update it (fiddling by hand) when we find new values. These files go in a hand/ directory at the same level as src/.
Similarly, it is often valuable to associate a constant value with a name:
conflict_begin: '2011-03-10'
So that we can refer in code to conflict_begin instead of hard-coding the date. This has several benefits: all references to this date now resolve to the same value, avoiding errors we might introduce if we mistyped the hard-coded date in several different places in the code. This also means that if we decide to change the value (perhaps we decide that the conflict started on 11 March instead of 10 March), we only have to change it once (don’t repeat yourself!). We say what we mean (“the start of the conflict”) instead of requiring that the person reading the code know what the date implies. We store data like this in yaml files in hand/.
frozen: In extremely rare cases, the best way to process a file is by using an interactive tool. For example, ancient files in dbf format (from dBase III, FoxBase, and FoxPro) can be easily and accurately read by LibreOffice — including the memo fields! — while other, scriptable tools tend to be buggy or partial. When we need to extract dbf files, we put the original dbf files in input/, then open them with LibreOffice, save the csv files to frozen/, and symlink the files from frozen/ to output/.